Project J3-4516
Neoantigens in non small cell lung cancer
Research project ARIS
Code: J3-4516
Period 1.10.2022 - 30.9.2025
Head: Assist Julij Šelb, MD, PhD
ARRS classification: Medicine/Human reproduction
Organisations: University Clinic of Respiratory and Allergic Diseases Golnik; National Institute of Biology
Description of the project
Efficient antitumor immunity in humans is to a large extent attributable to T-cells directed against neoantigens, present on tumor cells. Neoantigens are a group of HLA bound proteins, which are formed because of tumor specific mutations. Since they bypass central thymic tolerance they have high immunogenic potenitial. Therefore, they represent an attractive target for anti-tumor immunity. Pulmonary (lung) cancer is among the most frequent and by far the most deadly cancer type. Since smoking is a major risk factor for developing lung cancer it is, together with other cancers subjected to high carcinogen burden, also among the most somatic mutation (and probably also neoantigen) loaded tumor types.
The concept of neoantigens has successfully been used in routine clinical management of pulmonary and other cancers, mainly when treating those patients with immune checkpoint inhibitors (ICIs). Recently, FDA has approved treatment with ICI pembrolizumab in individuals with advanced tumors that have high tumor mutational burden (TMB; ≥10 mutations/megabase). TMB represents a proxy for evaluating neoantigen burden, however it is much less specific, since evaluation of TMB does not take into account all the biological steps needed for the presentation of the mutated DNA sequence as a neoantigen to the immune system.
ICIs work only on a subset of patients; in an unselected non small cell lung cancer (NSCLC) cohort, only around 20% of individuals are treatment responders. Since ICI have high burden of side effects (according to some estimations up to 60% of patients taking them experience side effects) and are also expensive medications it is therefore of paramount importance that only treatment responders get the treatment. However, biomarkers of treatment response are lacking. To date, solely the above mentioned TMB and immunohistochemical (IHC) expression of PD-L1 in the tumor tissue have been approved as clinical markers for guiding ICI therapy.
Different multiparametric prediction models (taking into account multiple tumor and host variables) have regularly shown improved prediction of ICI treatment response compared to only-TMB/only-PD-L1 IHC. Consistently, in these models, the most important prediction feature was TMB. In accordance with biological reasoning, a recent study has shown that solely neoantigen load (mutations presented and recognized by the immune system) is a better predictor of ICI treatment response than solely TMB. We hypothesize that using neoantigen load in such a context will significantly improve prediction accuracy of such a model; this will be a central hypothesis of the current project.
Therefore, in the project, we plan to set up neoantigen prediction pipelines, vi-vitro validate the result of those pipelines and use these results (neoantigen load) to refine ICI treatment response prediction models. Furthermore, we will evaluate the placement of the whole concept of using neoantigen refined ICI treatment response prediction procedure into local routine clinical practice - to see if patients and local health care system can benefit from it.
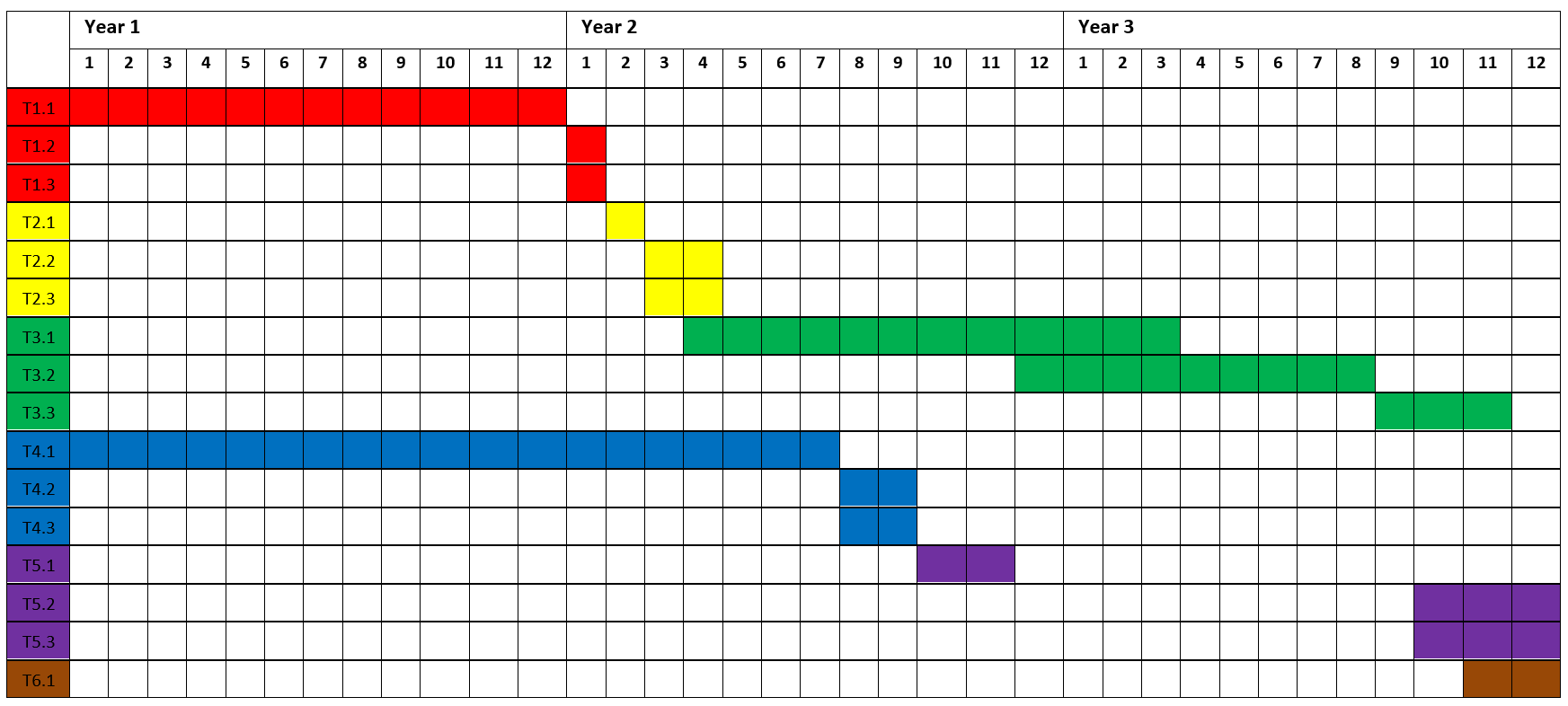
Project timeline
|
Work package (WP) |
Tasks (T) |
Milestones and deliverables |
Work distribution |
|
WP1 Sampling and sequencing (DNA, RNA) |
T1.1 Sampling of patients |
50 eligible patients (advanced NSLCL, 2 samples per tumor – central mass and tumorous lymph node) before starting treatment with ICI will be sampled |
GUC |
|
T1.2 WES of tumor samples |
100 WES of tumor samples will be performed (2 per patient) |
GUC |
|
|
T1.3 RNA seq of tumor samples (central tumor mass) |
50 RNA sequences of tumor sample (central mass) will be performed |
GUC |
|
|
WP2 Setting up established neoantigen burden/ICI response prediction algorithms |
T2.1 Setting up somatic mutation calling pipeline |
Somatic mutation calling pipeline will be up and running on GUC servers |
GUC |
|
T2.2 Setting up algorithms to establish neoantigen burden |
Neoantigen burden evaluating algorithms will be up and running on GUC servers |
GUC |
|
|
T2.3 Setting up ICI treatmen response prediction algorithms |
ICI treatment response prediction algorithms will be up and running on GUC servers |
GUC |
|
|
WP3 Experimental validation of prioritized neoantigen candidates |
T3.1 Synthesis of priority neoantigen peptides |
Predicted ‘presented and recognized’ peptides (neoantigens) of individuals with HLA-A*02:01 allele will be produced as nona/decamers |
ICI-NIB |
|
T3.2 Isolation/enrichment of CD8+ T cells from PBMCs |
Production of enriched neoantigen reactive CD8+ T cells required for further analyses |
ICI-NIB |
|
|
T3.3 Evaluation of reactivity against neoantigens |
Reactivity composite score for each evaluated neoantigen peptide will be calculated based on in vitro experiments |
ICI-NIB |
|
|
WP4 Evaluation of the established ICI treatment response prediction algorithms on the current cohort |
T4.1 Treatment of patients with ICIs |
Treatment of patients according to good clinical practice |
GUC |
|
T4.2 Defining responders/non responders |
Patients will be classified as responders/non-responders according to RECIST after 6 months of treatment |
GUC |
|
|
T4.3 Assessment of the established ICI response prediction algorithms |
ROC-AUC for differentiation between responders/non-responders with regard to established ICI response prediction algorithms will be produced |
GUC |
|
|
WP5 Refinement of the ICI treatment response prediction models using the project’s cohort data |
T5.1 Training of the existing algorithms on the project’s cohort data |
Models using existing predictors will be fitted to the data of the project’s cohort |
GUC |
|
T5.2 Refinement of the existing models with additional biologically plausible predictors |
Refined ICI prediction model will be trained on the project’s cohort data |
GUC |
|
|
T5.3 Comparing the models between each other |
List of ICI prediction models sorted according to ROC-AUC values will be produced |
GUC |
|
|
WP6 Pharmacoeconomical evaluation of the usage of best ICI prediction model in routine practice in the local environment |
T6.1 Pharmacoeconomical evaluation of the usage of best ICI prediction model in routine practice in the local environment |
Pharmacoeconomical estimation of incorporating best ICI response prediction algorithm to local routine practice will be produced |
GUC |
*GUC – Golnik university clinic; ICI-NIB - Immunology and Cellular Immunotherapy group at the National Institute of Biology
Project work packages and their realization


 04 25 69 100
04 25 69 100